8 min to read
基于bibliometrix进行文献分析
数据获取与转换
-
在pubmed中搜索
circular RNA,依次点击Send to > File > MEDLINE > Create File。 -
新建EndNote数据库,依次点击Edit > Output Styles > Open Style Manager > 勾选BibTeX Export,关闭窗口。即可输出BibTeX格式文件。
-
导入第一步下载的文件,依次点击File > Import > Import option > Other Filters >Pubmed(NLM) > 选择需要导入的文件 > import。
-
以BibTeX格式导出文献,选中需要导出的文件,依次点击File > Export > Output style: BibTex Export > 保存类型:Text File (*.txt)。
经测试,此方法可以大规模获取BibTeX格式数据,供后续分析。
实操
数据整理
import os
import re
os.chdir("C:\\Users\\Administrator\\Desktop")
with open('pubmed_result.txt', 'r') as f:
str=f.read()
str01=re.split(r'([A-Z]{2,4}[ ]{0,2}-[ ]{1})',str) #正则表达式外加小括号表示保留匹配的字符,大括号表示匹配次数,大括号内2,4表示匹配2-4次
library(bibliometrix)
D <- readFiles("http://www.bibliometrix.org/datasets/savedrecs.bib")
M <- convert2df(D, dbsource = "isi", format = "bibtex")
#初步分析
results <- biblioAnalysis(M, sep = ";")
options(width=100)
S <- summary(object = results, k = 10, pause = FALSE)
plot(x = results, k = 10, pause = FALSE)
#引用分析(Analysis of Cited References)
CR <- citations(M, field = "article", sep = ". ")
cbind(CR$Cited[1:10])
CR <- citations(M, field = "author", sep = ". ")
cbind(CR$Cited[1:10])
CR <- localCitations(M, sep = ". ")
CR$Authors[1:10,]
CR$Papers[1:10,]
#Authors’ Dominance ranking
DF <- dominance(results, k = 10)
#Authors’ h-index
indices <- Hindex(M, authors="BORNMANN L", sep = ";",years=10)
indices$H
indices$CitationList
authors=gsub(","," ",names(results$Authors)[1:10])
indices <- Hindex(M, authors, sep = ";",years=50)
indices$H
#Lotka’s Law coefficient estimation
L <- lotka(results)
L$AuthorProd
Observed=L$AuthorProd[,3]
# Theoretical distribution with Beta = 2
Theoretical=10^(log10(L$C)-2*log10(L$AuthorProd[,1]))
plot(L$AuthorProd[,1],Theoretical,type="l",col="red",ylim=c(0, 1), xlab="Articles",ylab="Freq. of Authors",main="Scientific Productivity")
lines(L$AuthorProd[,1],Observed,col="blue")
legend(x="topright",c("Theoretical (B=2)","Observed"),col=c("red","blue"),lty = c(1,1,1),cex=0.6,bty="n")
#Bipartite networks
A <- cocMatrix(M, Field = "SO", sep = ";")
sort(Matrix::colSums(A), decreasing = TRUE)[1:5]
# A <- cocMatrix(M, Field = "CR", sep = ". ") #Citation network
# A <- cocMatrix(M, Field = "AU", sep = ";") #Author network
# M <- metaTagExtraction(M, Field = "AU_CO", sep = ";") #Country network
# A <- cocMatrix(M, Field = "AU_CO", sep = ";") #Country network
#Bibliographic coupling
NetMatrix <- biblioNetwork(M, analysis = "coupling", network = "authors", sep = ";")
net=networkPlot(NetMatrix, normalize = "salton", weighted=NULL, n = 100, Title = "Authors' Coupling", type = "fruchterman", size=5,size.cex=T,remove.multiple=TRUE,labelsize=0.8,label.n=10,label.cex=F)
#Bibliographic collaboration
NetMatrix <- biblioNetwork(M, analysis = "collaboration", network = "authors", sep = ";")
#Descriptive analysis of network graph characteristics
NetMatrix <- biblioNetwork(M, analysis = "co-occurrences", network = "keywords", sep = ";")
netstat <- networkStat(NetMatrix)
names(netstat$network)
summary(netstat, k=10)
#Visualizing bibliographic networks
M <- metaTagExtraction(M, Field = "AU_CO", sep = ";")
NetMatrix <- biblioNetwork(M, analysis = "collaboration", network = "countries", sep = ";")
net=networkPlot(NetMatrix, n = dim(NetMatrix)[1], Title = "Country Collaboration", type = "circle", size=TRUE, remove.multiple=FALSE,labelsize=0.7,cluster="none")
NetMatrix <- biblioNetwork(M, analysis = "co-occurrences", network = "keywords", sep = ";")
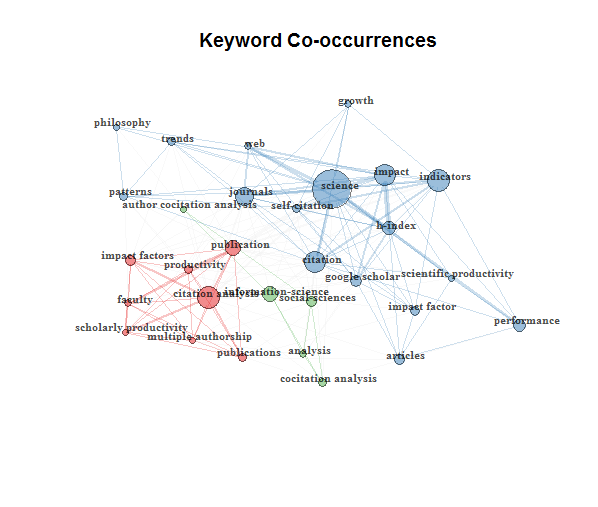
net=networkPlot(NetMatrix, normalize="association", weighted=T, n = 30, Title = "Keyword Co-occurrences", type = "fruchterman", size=T,edgesize = 5,labelsize=0.7)
#Co-Word Analysis
CS <- conceptualStructure(M,field="ID", method="CA", minDegree=4, k.max=8, stemming=FALSE, labelsize=10, documents=10)
#Historical Direct Citation Network
options(width=130)
histResults <- histNetwork(M, min.citations = 10, sep = ". ")
net <- histPlot(histResults, n=10, size = 10, labelsize=5, size.cex=TRUE, arrowsize = 0.5, color = TRUE)
参考资料
[01]. A brief introduction to bibliometrix
[02]. 文献阅读的文本分析流派

Comments